Research
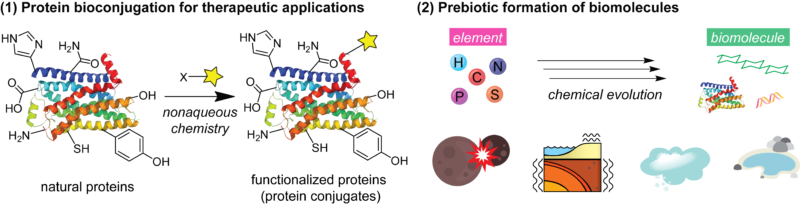
Selective and efficient reactions of polypeptides in living systems are enabled by a range of enzymes and molecular machinery even in a complex mixture of cellular components. During the past decades, nonenzymatic reactions or chemical modification of polypeptides have been extensively studied by addressing challenges of aqueous chemical transformation of the polyfunctional biomolecules for various purposes such as creation of therapeutic agents as well as investigation of biomolecular processes. Nonetheless, realization of selective, effective chemical reactions of polypeptides in aqueous media still represents a formidable challenge. Recognizing the existence of nonaqueous environments where polypeptides would not necessarily lose their structural integrity and functions, the central research theme of the Ohata group is to examine chemical reactivities of polypeptides in nonaqueous media. One aspect of the research campaign is to achieve nonaqueous bioconjugation for creation of therapeutic agents such as antibody–drug conjugates. This group also has another program about chemical reactivities of polypeptides in prebiotically plausible conditions such as those in the early Earth and other astrochemical events.
In addition, this is not about our ongoing research, but we also developed an equitable learning tool by which you can improve your science writing with free resources. Check it out! https://pubs.acs.org/doi/10.1021/acs.jchemed.4c00781